Researchers have identified five types of viruses that can cause illness in humans or livestock. Notably, one type is closely related to SARS-CoV-2 and SARS.
According to The Telegraph, a new study has discovered a novel coronavirus-like virus identified in bats in southern China. Chinese and Australian scientists sampled 149 bats across Yunnan Province, which borders Laos and Myanmar. As a result, they identified five types of viruses “capable of causing disease in humans or livestock.” Among these, one type of bat coronavirus is closely related to both SARS and SARS-CoV-2.

A researcher from the Franceville International Medical Research Center collects bats using a net on November 25, 2020, inside a cave in Gabon. (Photo: Steeve Jordan/AFP).
Recombinant Viruses Forming New Pathogens
Professor Eddie Holmes, an evolutionary biologist and virologist at the University of Sydney and co-author of the study, stated that this finding indicates that SARS-CoV-2-like viruses continue to circulate among bats and pose ongoing risks.
The study, published as a preprint and awaiting peer review, revealed that bats are often infected with multiple types of viruses simultaneously. This information is crucial as it shows the potential for current viruses to exchange genetic material (or recombine) to form new pathogens.
Virologist Professor Jonathan Ball from the University of Nottingham, who did not participate in the study, commented: “Each bat can harbor many different species of viruses, sometimes becoming a host to multiple types simultaneously. Co-infection, especially with related viruses like coronaviruses, creates opportunities for viruses to exchange important genetic information, resulting in new natural variants.”
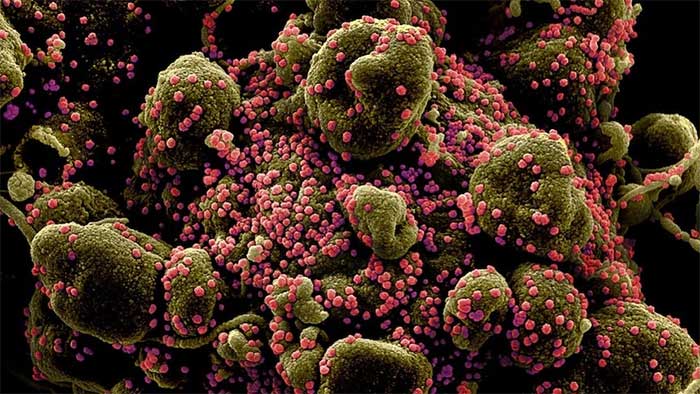
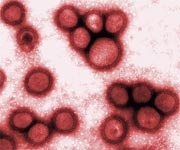
A dying cell (brownish-green) infected with SARS-CoV-2 virus particles (pink). This image was isolated from a sample provided by the National Institute of Allergy and Infectious Diseases in the U.S. (Photo: NIH).
Clear Threats
Professor Stuart Neil, Head of the Department of Infectious Diseases at King’s College London, added: “This study provides us with very important insights into the evolution and ecosystem of coronaviruses, the extent to which they recombine, and frequently transition into new species.”
The expert emphasized that this also serves as evidence of the clear and present threat of new disease outbreaks in humans.
Previously, an analysis estimated that up to 400,000 people are infected by bat viruses each year across southern China and Southeast Asia.
Among the five viruses labeled “of concern,” one possesses characteristics of both SARS and SARS-CoV-2. SARS is the disease that killed 774 people and infected over 8,000 during an outbreak in 2003. Meanwhile, the death toll from SARS-CoV-2 has reached tens of millions.
This virus has been named BtSY2.
Notably, BtSY2 has a receptor-binding domain very similar to that of nCoV. The receptor-binding domain is part of the spike protein, which the virus uses to attach to human cells. This is the target that most Covid-19 vaccines are currently aimed at.
BtSY2 is also the most similar to nCoV ever identified. This suggests that BtSY2 could also infect humans.
Professor Holmes noted: “It is closely related to the BANAL bat virus from Laos and another virus we have previously seen from China.”
The new study does not explain how SARS-CoV-2 initially jumped to humans, nor does it rule out a laboratory accident leading to the virus’s leak. However, it aids scientists in tracking the potential evolutionary process of the virus.
This research follows a new analysis presented at the Health Congress in Singapore earlier in November, which suggested that some bat coronaviruses share a common ancestor with SARS-CoV-2 dating back to 2016. They based this on comparative results of viral gene segments.
Professor Joel Wertheim, an evolutionary biologist at the University of California, San Diego, and co-author, stated: “We need to sequence the entire viral genome of the circulating bat viruses, not just small fragments of it. If we don’t assemble the small fragments of this viral genome, we may miss important segments that reveal the history of SARS-CoV-2.”