Scientists at Meta, the parent company of Facebook and Instagram, have utilized a language model powered by artificial intelligence (AI) to predict the unknown structures of over 600 million proteins from viruses, bacteria, and other microorganisms.
The program, named ESMFold, employed an initial model designed to decode human language to make accurate predictions about the folds created by proteins that determine their 3D structures.
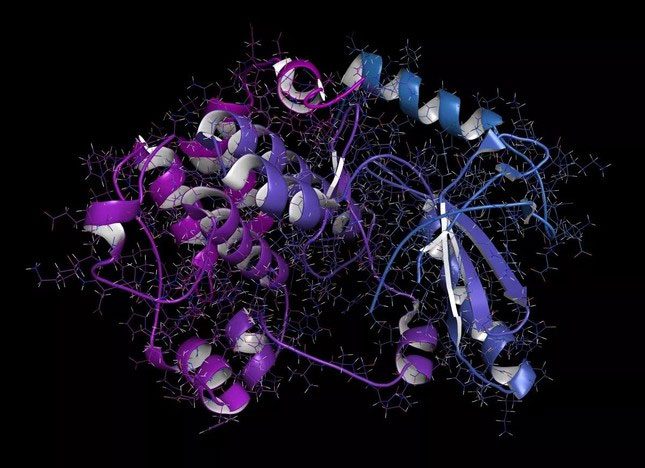
A MEK1 or mitogen-activated protein kinase 1 (rabbit) activated by mitogen.
These predictions, compiled in the ESM Metagenomic Atlas, an open-source resource, can be utilized to aid in the development of new drugs, describe the functions of previously unknown microorganisms, and track evolutionary relationships among distantly related species.
ESMFold is not the first program to make predictions about proteins. In 2022, DeepMind, a subsidiary of Google, announced that its protein prediction program AlphaFold had decoded the shapes of approximately 200 million proteins known to science.
Meta stated that while ESMFold is not as accurate as AlphaFold, it is 60 times faster than DeepMind’s program. The results have not yet been peer-reviewed.
Knowing the shape of a protein is the best way to understand its function, but there are astonishing ways that combinations of amino acids in different sequences can form.
The gold standard for determining protein structure is using X-ray crystallography—observing how high-energy light beams diffract around proteins—but this is a labor-intensive method that can take months or years to yield results and is not efficient for all types of proteins. After decades of work, over 100,000 protein structures have been deciphered through X-ray crystallography.
To tackle this problem, Meta researchers turned to a complex computer model designed to decode and make predictions about human language, applying this model to the language of protein sequences.
To test this model, scientists accessed databases of DNA and genes sourced from diverse environments such as soil, seawater, human intestines, and skin. By inputting DNA data into the ESMFold program, researchers predicted the structures of over 617 million proteins in just two weeks.
This figure exceeds AlphaFold’s claim of having decoded the structures of around 200 million proteins four months ago, when they announced they had inferred the protein structures of nearly every known type.
This implies that many of these proteins have never been seen before, possibly because they originate from unknown organisms. Over 200 million predictions made by ESMFold are believed to be of high quality, indicating that the program was able to predict shapes with atomic-level accuracy.