With the advancement of science and technology, AI can be utilized to discover new treatment methods and high-tech biomaterials.
The recent resurgence of artificial intelligence (AI) following the collapse of the 1980s – known as the “AI winter” – has proven to be truly revolutionary. Recently, DeepMind, a subsidiary of Google tasked with developing advanced AI programs, revealed that their AlphaFold system has decoded the structure of nearly all known proteins – over 200 million of them.
At this point in time, a new AI tool is pushing the boundaries of what is possible even further by allowing scientists to design original proteins that are unlike anything found in nature. This new tool, known as ProteinMPNN, has recently been described by researchers at the University of Washington in a pair of studies published in the journal Science.
The authors believe that ProteinMPNN and other similar tools linked to the surface in the near future will open up a new realm of possibilities and subsequent applications. These include entirely new proteins designed from scratch to meet specific goals, which could be digestive enzymes for plastics or new drugs targeting some of the most chronic and challenging diseases today.
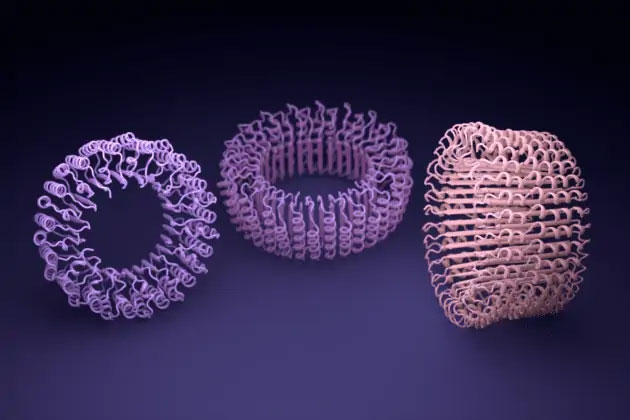
Proteins are essential for cell life. They perform complex tasks and act as catalysts for chemical reactions. For a long time, many scientists have sought to harness this power by designing artificial proteins that can perform new functions such as treating diseases, capturing carbon, or storing energy. However, many processes for creating such proteins are quite slow due to their complexity and high failure rates. Previous studies have generated many methods to shape protein structures, but capturing their functions has proven difficult.
The role of proteins in supporting life and nature as a whole is immensely significant. Some are structural, others transport molecules, and some serve as receptors, among other functions. Each of these functions is closely related to its specific shape.
All proteins begin as a linear chain of basic units known as amino acids. The main 2D structure of these amino acids contains the “recipe” that proteins use to fold themselves. A protein goes through repeated folding stages, applying a series of configurations before reaching its final shape, which is the most energetically favorable configuration.
While AlphaFold can predict the shapes of existing proteins and infer their functions, ProteinMPNN can tackle a similar problem but from the reverse angle. Instead of reverse-engineering the roles of proteins from nature, the new tool can help scientists design entirely new proteins from scratch. For instance, they can design a specific function or purpose for a protein and then use AI to generate the corresponding structure that molecular components and shapes would favor the desired function. The remaining challenge is to synthesize these proteins in the laboratory.
ProteinMPNN achieves all these remarkable feats by utilizing two powerful AIs developed at the University of Washington. The first tool, named “hallucination”, allows scientists to search among potential useful protein sequences based on simple prompts – similar to the famous AI image generator DALL-E, which creates stunning images from text prompts.
The second AI, known as “inpainting”, can be seen as an autocomplete feature like the one you experience when typing a query into Google – but specifically for proteins. When used in synergy, these two AIs can enable scientists to discover entirely new proteins that fit the desired function.
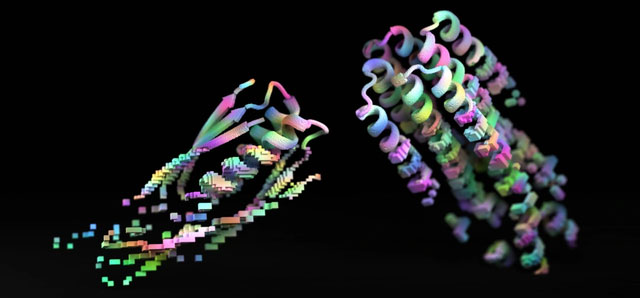
By developing machine learning models capable of reviewing selected protein information from gene databases, researchers have uncovered relatively simple design rules for creating new artificial proteins. When constructing these proteins in physical laboratories, they found that they performed chemically well enough to compete with those available in nature.
To validate the various protein shapes generated by the two AIs, researchers transferred the data to AlphaFold and tested whether the amino acid sequences could indeed fold into the desired shapes.
The initially designed proteins using ProteinMPNN were then assembled in the laboratory. Among them were structures only nanometers in size, which could be fitted inside custom nano devices.
“This is the starting point for machine learning in protein design. In the coming months, we will work to improve these tools to create even more dynamic and functional proteins,” said senior author David Baker, a professor of biochemistry at the University of Washington School of Medicine.